Lecture 04-2: Data Science at the Command Line
DATA 503: Fundamentals of Data Engineering
February 2, 2026
Introduction to Data Science at the Command Line
The command line has been around for over 50 years, yet it remains one of the most powerful tools for CS, DS, and beyond! Today we explore why.
Why the Command Line?
Data Science Meets Ancient Technology
How can a technology that is more than 50 years old be useful for a field that is only a few years young?
Today, data scientists can choose from an overwhelming collection of exciting technologies:
- Python, R, Julia
- Apache Spark
- Jupyter Notebooks
- GUI-based tools
And yet, the command line remains essential. Let us understand why.
Five Advantages of the Command Line
The command line offers five key advantages for data science:
| Advantage | Description |
|---|---|
| Agile | REPL environment allows rapid iteration and exploration |
| Augmenting | Integrates with and amplifies other technologies |
| Scalable | Automate, parallelize, and run on remote servers |
| Extensible | Create your own tools in any language |
| Ubiquitous | Available on virtually every system |
The Command Line Is Agile
Data science is interactive and exploratory. The command line supports this through:
Read-Eval-Print Loop (REPL)
- Type a command, press Enter, see results immediately
- Stop execution at will
- Modify and re-run quickly
- Short iteration cycles let you play with your data
Close to the Filesystem
- Data lives in files
- Command line offers convenient tools for file manipulation
- Navigate, search, and transform data files easily
The Command Line Is Augmenting
The command line does not replace your current workflow. It amplifies it.
Three ways it augments your tools:
- Glue between tools: Connect output of one tool to input of another using pipes
- Delegation: Python, R, and Spark can call command-line tools and capture output
- Tool creation: Convert your scripts into reusable command-line tools
Every technology has strengths and weaknesses. Knowing multiple technologies lets you use the right one for each task.
The Command Line Is Scalable
On the command line, you do things by typing. In a GUI, you point and click.
Everything you type can be:
- Saved to scripts
- Rerun when data changes
- Scheduled to run at intervals
- Executed on remote servers
- Parallelized across multiple cores
Pointing and clicking is hard to automate. Typing is not.
The Command Line Is Extensible
The command line itself is over 50 years old, but its tools are being developed daily.
- Language agnostic: tools can be written in any programming language
- Open source community produces high-quality tools
- Tools can work together
- You can create your own tools
The Command Line Is Ubiquitous
The command line comes with any Unix-like operating system:
- macOS
- Linux (100% of top 500 supercomputers)
- Windows Subsystem for Linux
- Cloud servers
- Embedded systems
This technology has been around for more than five decades. It will be here for five more.
The OSEMN Model
Data Science Is OSEMN
The field of data science employs a practical definition devised by Hilary Mason and Chris H. Wiggins [1].
OSEMN (pronounced “awesome”) defines data science in five steps:
- Obtaining data
- Scrubbing data
- Exploring data
- Modeling data
- iNterpreting data
OSEMN: Expectation vs Reality
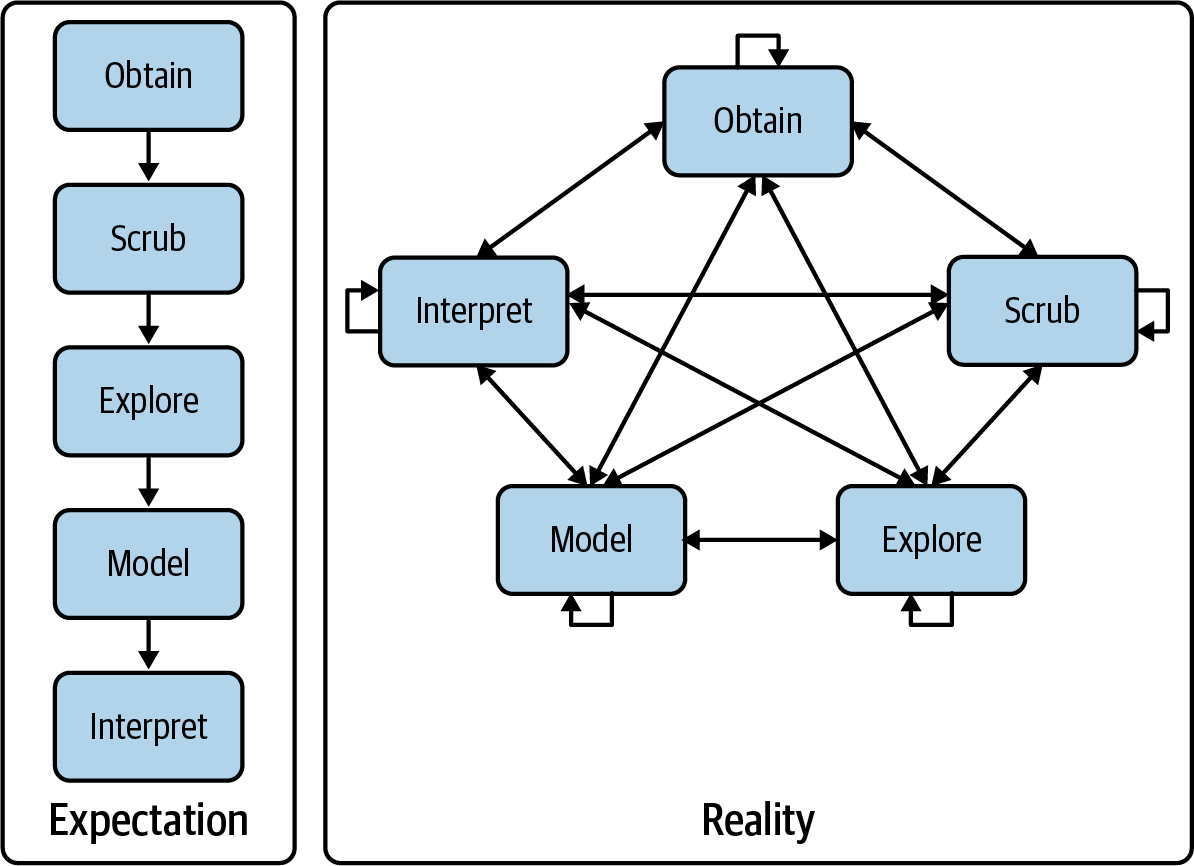
The OSEMN model shows data science as an iterative, nonlinear process
In practice, you move back and forth between steps. After modeling, you may return to scrubbing to adjust features.
The OSEMN Steps
Download, query APIs, extract from files, or generate data yourself.
Common formats: plain text, CSV, JSON, HTML, XML
Clean the data before analysis:
- Filter lines
- Extract columns
- Replace values
- Handle missing data
- Convert formats
80% of data project work is in cleaning the data.
Get to know your data:
- Look at the data
- Derive statistics
- Create visualizations
Create statistical models:
- Clustering
- Classification
- Regression
- Dimensionality reduction
Draw conclusions, evaluate results, and communicate findings.
This is where human judgment matters most.
Getting Started
Setting Up Your Environment
Docker: Your Portable Environment
We use Docker to ensure everyone has the same environment with all necessary tools.
What is Docker?
- A Docker image bundles applications with their dependencies
- A Docker container is an isolated environment running an image
- Like a lightweight virtual machine, but uses fewer resources
Why Docker for this course?
- Consistent environment across Windows, macOS, and Linux
- All tools pre-installed and configured
- No dependency conflicts
- Easy to reset if something goes wrong
Installing Docker
Download Docker Desktop from docker.com
Install and launch Docker Desktop
Open your terminal (or Command Prompt on Windows)
Pull the course Docker image:
Running the Docker Container
Run the Docker image:
You are now inside an isolated environment with all the necessary command-line tools installed.
Test that it works:
Mounting a Local Directory
To get data in and out of the container, mount a local directory:
The -v option maps your current directory to /data inside the container.
Exiting the Container
When you are done, exit the container by typing:
The container is removed (due to --rm flag), but any files you saved to the mounted directory persist on your computer.
What Is the Command Line?
The Terminal
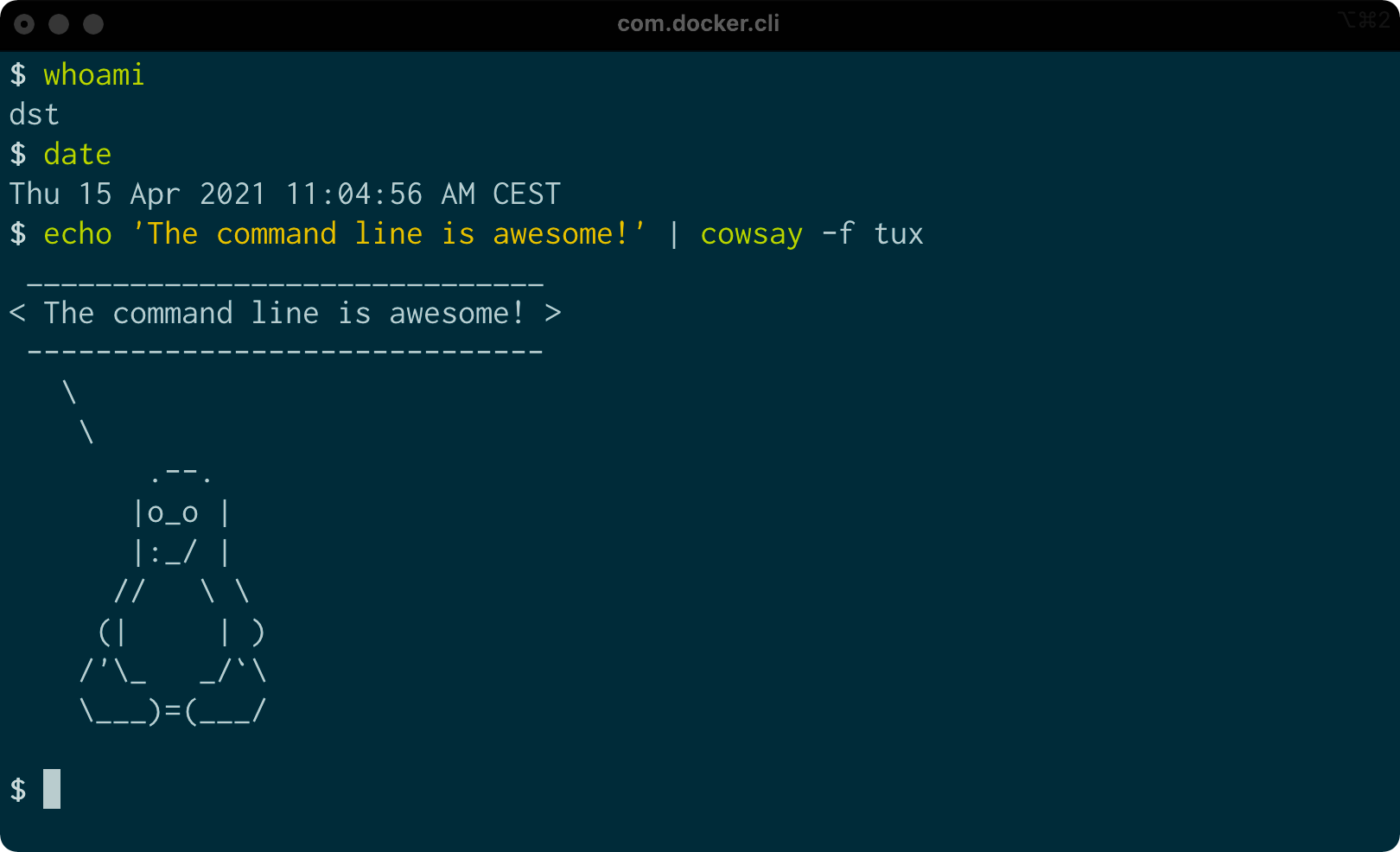
Command line on macOS
The terminal is the application where you type commands. It enables you to interact with the shell.
The Terminal on Linux
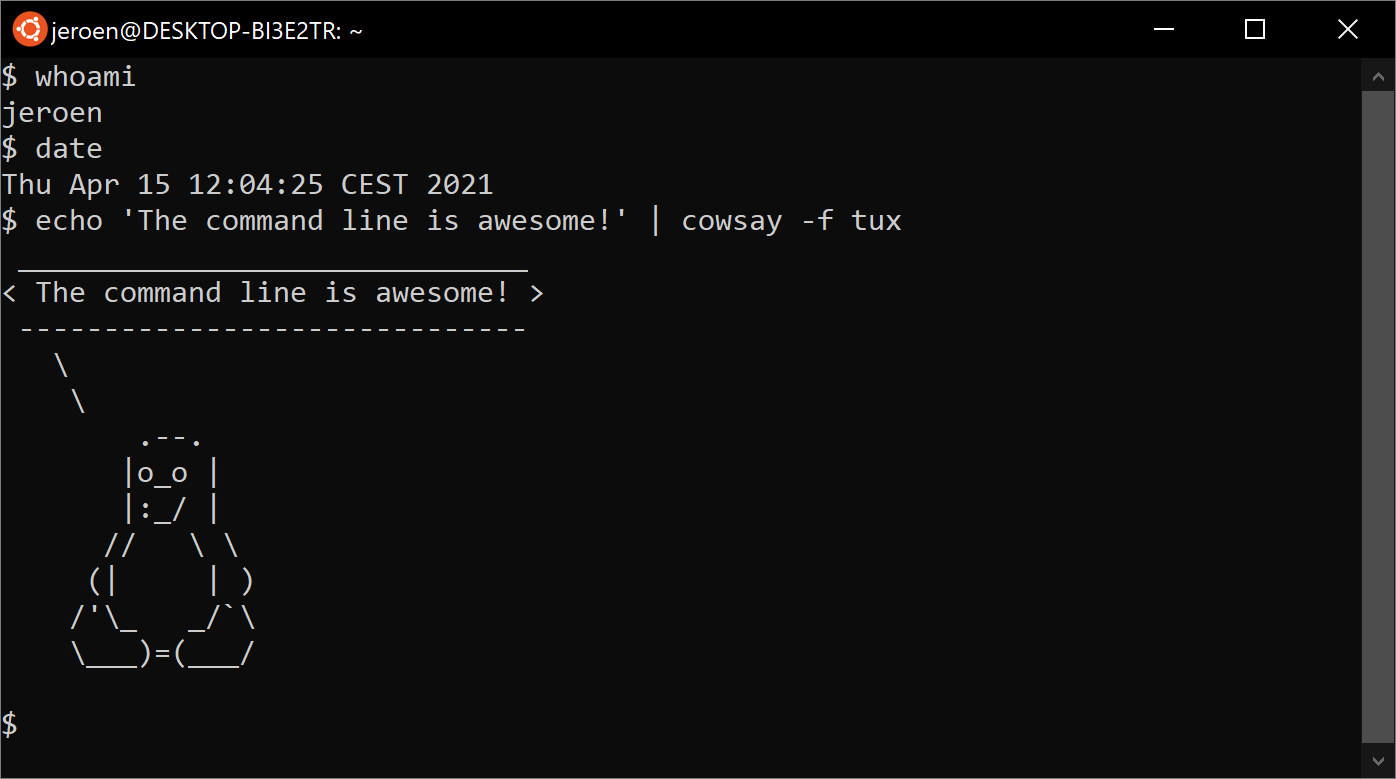
Command line on Ubuntu
Ubuntu is a distribution of GNU/Linux. The same commands work across different Unix-like systems.
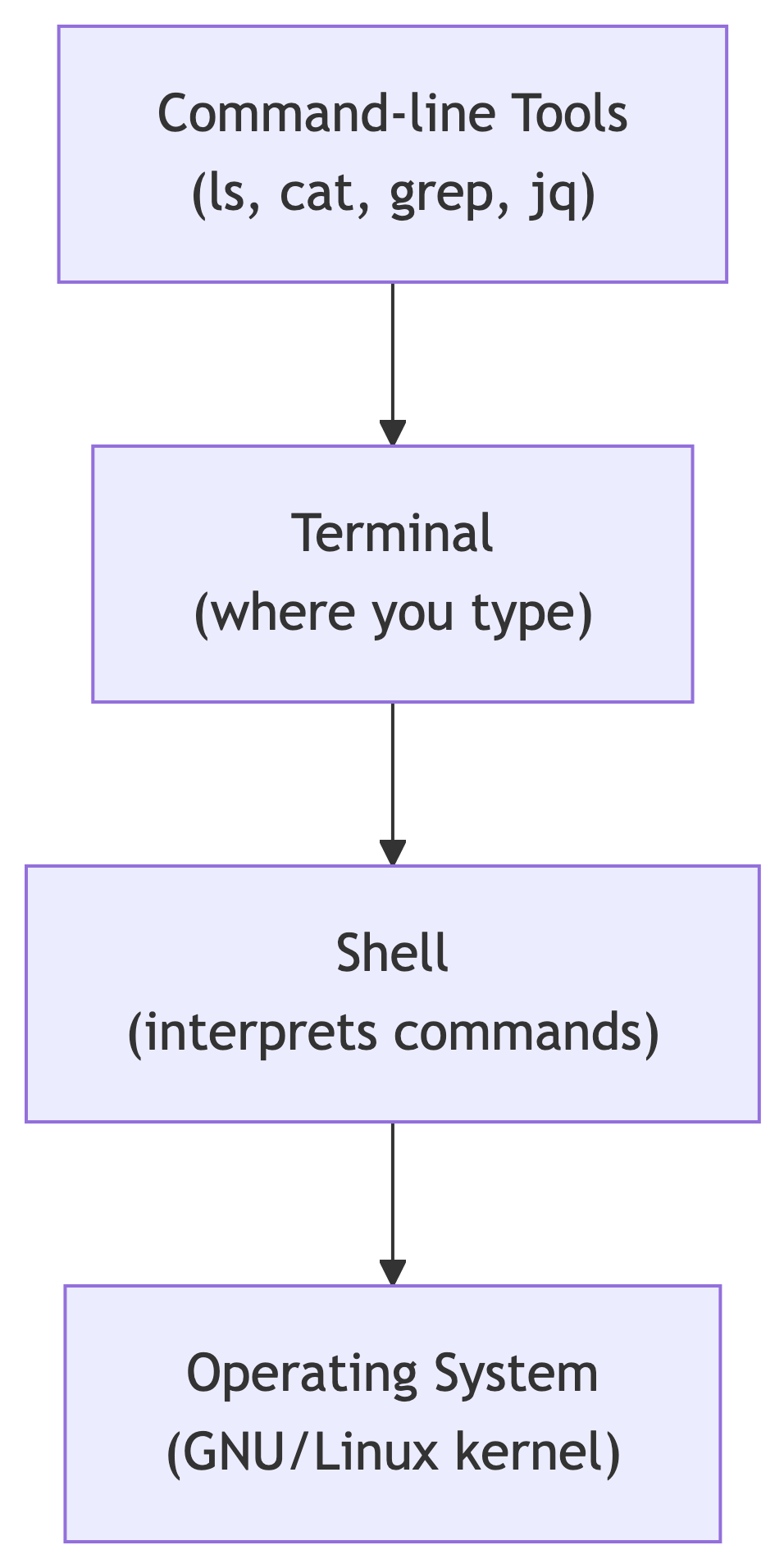
The Four Layers
The environment consists of four layers:
| Layer | Purpose |
|---|---|
| Command-line tools | Programs you execute |
| Terminal | Application for typing commands |
| Shell | Program that interprets commands (bash, zsh) |
| Operating system | Executes tools, manages hardware |
Understanding the Prompt
When you see text like this in the book or slides:
- The
$is the prompt (do not type it) seq 3is the command you type1,2,3is the output
The prompt may show additional information (username, directory, time), but we show only $ for simplicity.
Essential Unix Concepts
Executing Commands
Your First Commands
Try these commands in your terminal:
The tool pwd outputs the name of your current directory.
The tool cd changes directories. Values after the command are called arguments or options.
Command Arguments and Options
Commands often take arguments and options:
This command has three arguments:
| Argument | Type | Purpose |
|---|---|---|
-n |
Option (short form) | Specifies number of lines |
3 |
Value | The number of lines to show |
movies.txt |
Filename | The file to read |
The long form of -n is --lines.
Five Types of Command-Line Tools
The Tool Umbrella
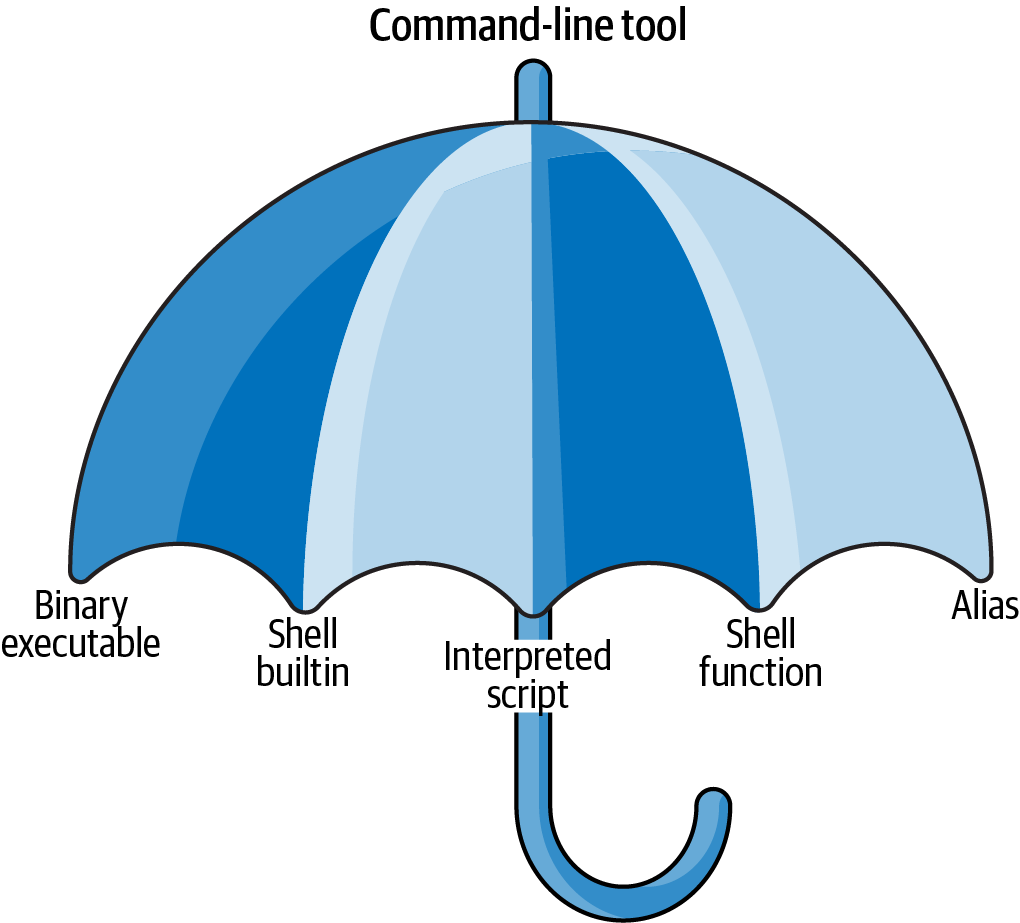
Each command-line tool is one of five types:
- Binary executable
- Shell builtin
- Interpreted script
- Shell function
- Alias
Binary Executables
Programs compiled from source code to machine code.
- Created by compiling C, C++, Rust, Go, etc.
- Cannot read the file in a text editor
- Fast execution
- Examples:
ls,grep,cat
Shell Builtins
Command-line tools provided by the shell itself.
- Part of the shell (bash, zsh)
- May differ between shells
- Cannot be easily inspected
- Examples:
cd,pwd,echo
Interpreted Scripts
Text files executed by an interpreter (Python, R, Bash).
Advantages: Readable, editable, portable
Shell Functions
Functions executed by the shell itself:
- Similar to scripts but typically smaller
- Defined in shell configuration (
.bashrc,.zshrc) - Good for personal productivity shortcuts
Aliases
Macros that expand to longer commands:
Now typing l expands to the full ls command with options.
- Save keystrokes for common commands
- Fix common typos
- Cannot take parameters (use functions for that)
Identifying Tool Types
Use the type command to identify what kind of tool you have:
Combining Tools
The Unix Philosophy
Most command-line tools follow the Unix philosophy:
Do one thing and do it well.
grepfilters lineswccounts lines, words, characterssortsorts linesheadshows first lines
The power comes from combining these simple tools.
Standard Streams
Every tool has three standard communication streams:
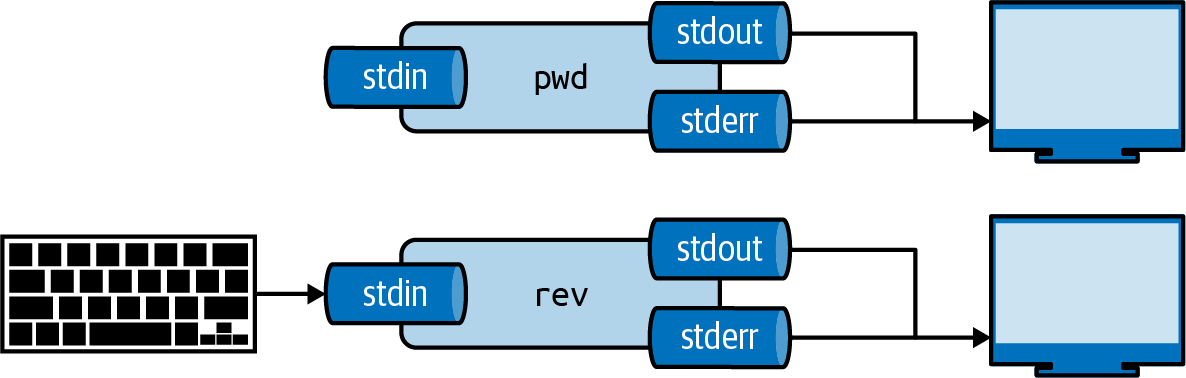
Standard input (stdin), standard output (stdout), and standard error (stderr)
| Stream | Abbreviation | Default |
|---|---|---|
| Standard input | stdin | Keyboard |
| Standard output | stdout | Terminal |
| Standard error | stderr | Terminal |
The Pipe Operator
The pipe operator (|) connects stdout of one tool to stdin of another:
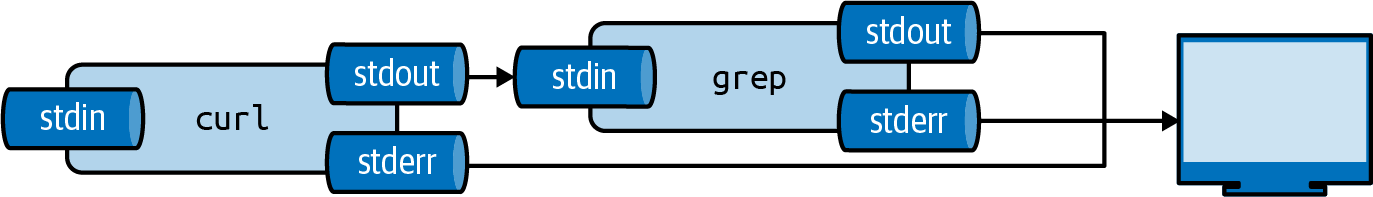
Piping output from curl to grep
Chaining Multiple Tools
You can chain as many tools as needed:
This pipeline:
- Downloads Alice in Wonderland
- Filters lines containing ” CHAPTER”
- Counts the number of lines
Think of piping as automated copy and paste.
Redirecting Input and Output
Output Redirection
Save output to a file using >:
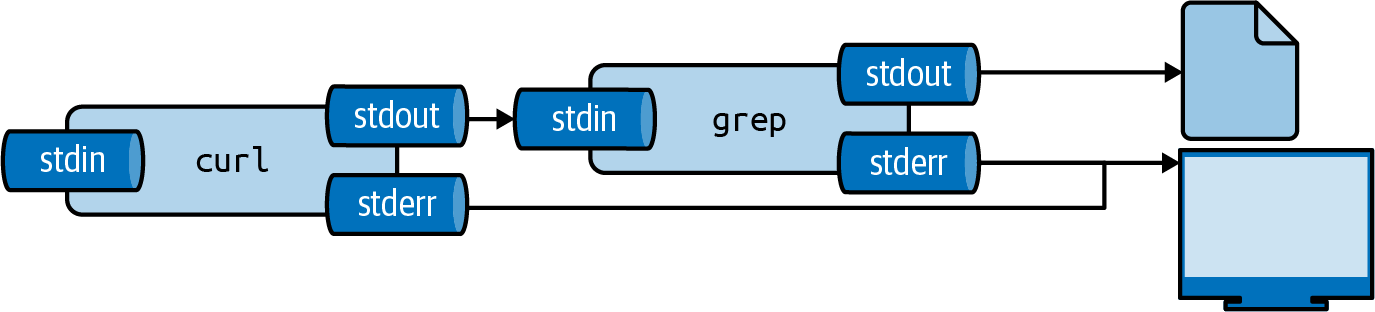
Redirecting output to a file
- Creates file if it does not exist
- Overwrites file if it exists
- Standard error still goes to terminal
Appending Output
Use >> to append instead of overwrite:
The -n option tells echo not to add a trailing newline.
Input Redirection
Read from a file using <:
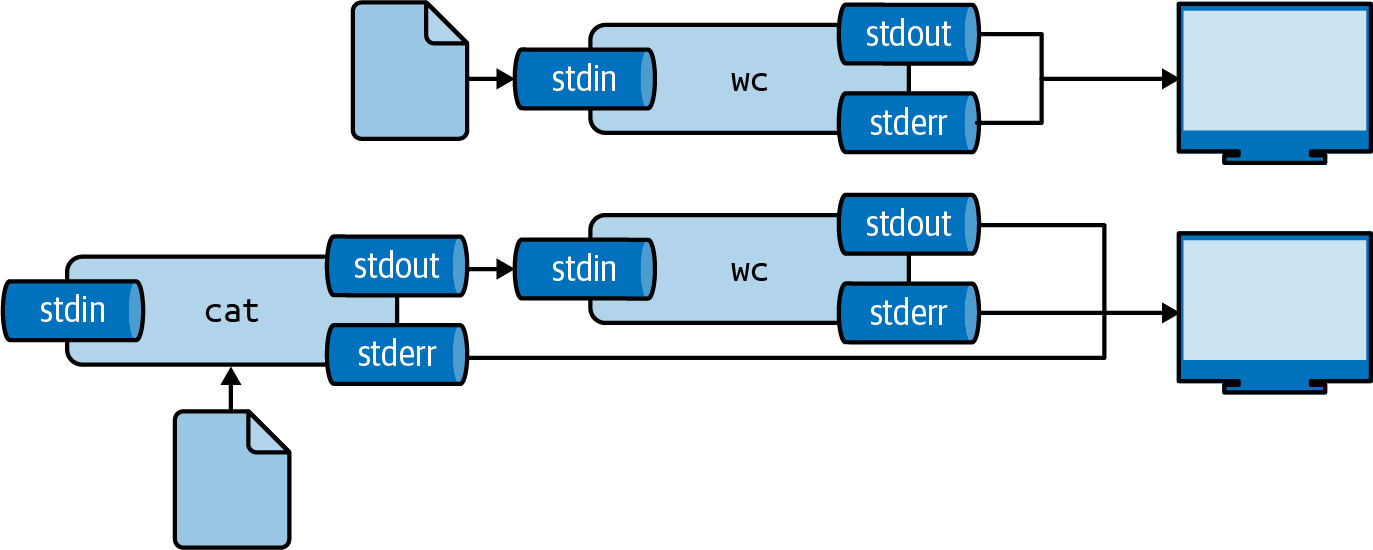
Two ways to use file contents as input
Both achieve the same result. The second form avoids starting an extra process.
Redirecting Standard Error
Suppress error messages by redirecting stderr to /dev/null:
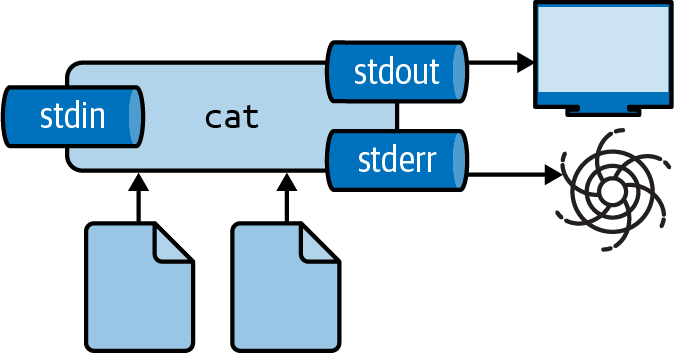
Redirecting stderr to /dev/null
The 2 refers to standard error (file descriptor 2).
The Sponge Problem
Warning: Do not read from and write to the same file in one command!
The output file is opened (and emptied) before reading starts.
Solutions:
- Write to a different file, then rename
- Use
spongeto absorb all input before writing
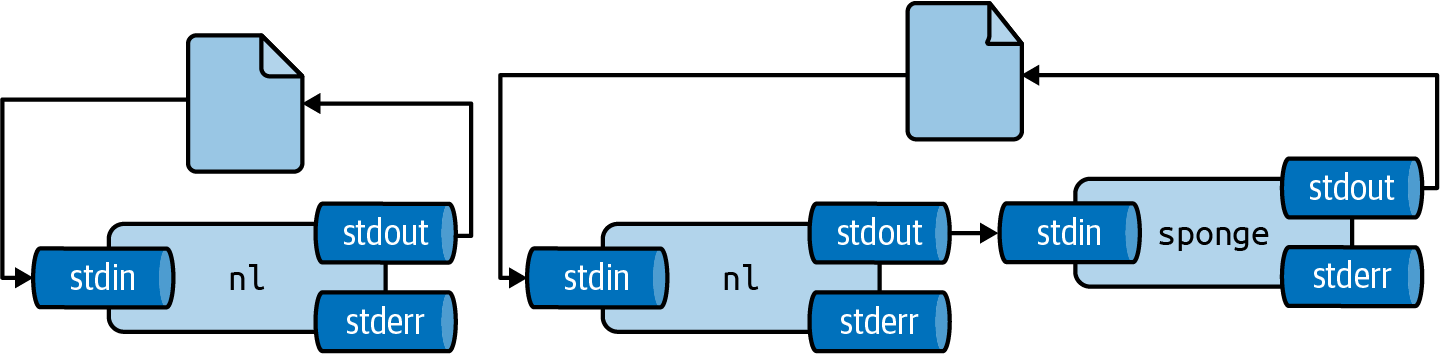
sponge soaks up input before writing
Using tee for Intermediate Output
The tee command writes to both a file and stdout:
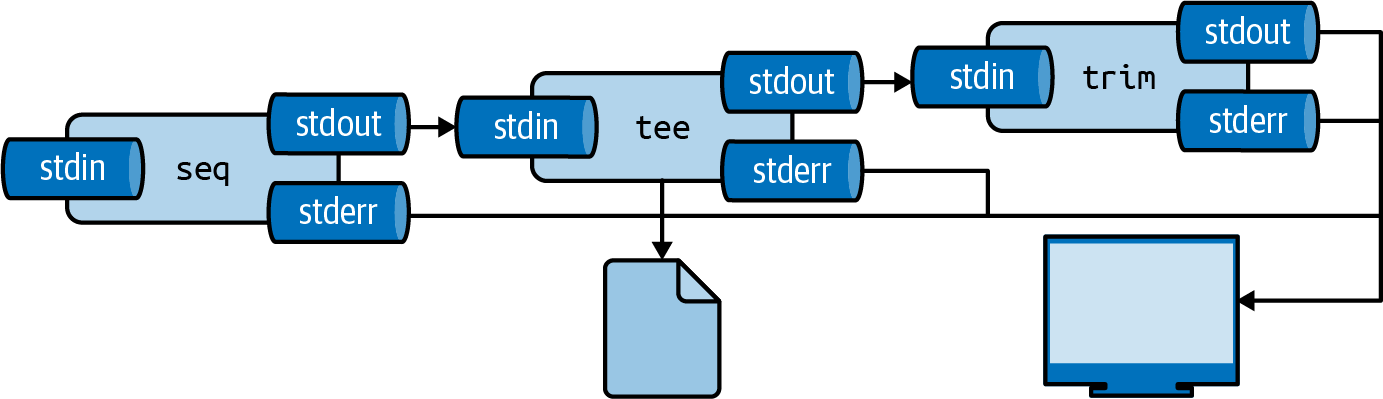
tee writes to file while passing data through
Useful for saving intermediate results while continuing a pipeline.
Working with Files and Directories
Listing Directory Contents
Common options:
| Option | Meaning |
|---|---|
-l |
Long format (permissions, size, date) |
-h |
Human-readable sizes |
-F |
Append indicators (/ for directories) |
-a |
Show hidden files |
Creating and Navigating Directories
Moving and Copying Files
Removing Files and Directories
Warning: There is no recycle bin on the command line. Deleted files are gone.
Use the -v (verbose) option to see what is happening:
Getting Help
Manual Pages
The man command displays the manual page for a tool:
Navigation:
- Space or
f: next page b: previous page/pattern: searchq: quit
The –help Option
Many tools support --help:
Some tools use -h instead.
TLDR Pages
For concise, example-focused help, use tldr:
Shows practical examples instead of exhaustive documentation.
Shell Builtins Help
For shell builtins like cd, check the shell manual:
Or use the help command (in bash):
Summary
Key Takeaways
What We Learned
OSEMN Model: Obtain, Scrub, Explore, Model, iNterpret - data science is iterative
Command Line Advantages: Agile, augmenting, scalable, extensible, ubiquitous
Docker Setup: Use
lucascordova/dataengfor a consistent environmentTool Types: Binary executables, shell builtins, scripts, functions, aliases
Pipes and Redirection: Combine simple tools into powerful pipelines
File Management: Navigate, create, copy, move, and remove files
Looking Ahead
Next steps:
- Practice the commands covered today
- Explore the Docker environment
- Complete the command-line exercises
Questions?
References
References
Mason, H. & Wiggins, C. H. (2010). A Taxonomy of Data Science. dataists blog. http://www.dataists.com/2010/09/a-taxonomy-of-data-science
Janssens, J. (2021). Data Science at the Command Line (2nd ed.). O’Reilly Media. https://datascienceatthecommandline.com
Raymond, E. S. (2003). The Art of Unix Programming. Addison-Wesley.
Patil, D. J. (2012). Data Jujitsu. O’Reilly Media.
Docker Documentation. https://docs.docker.com
GNU Coreutils Manual. https://www.gnu.org/software/coreutils/manual